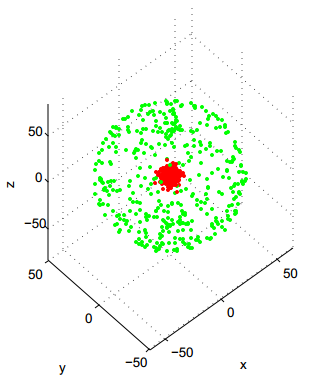
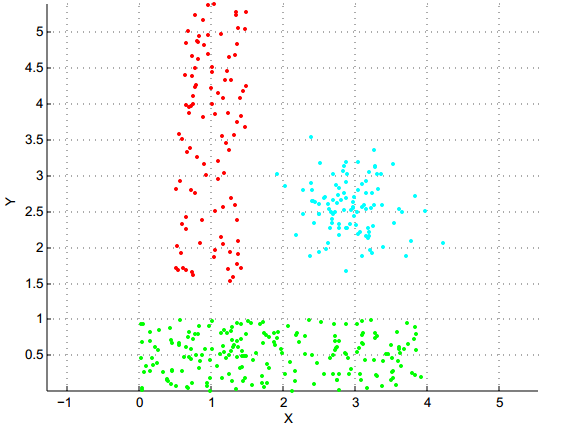
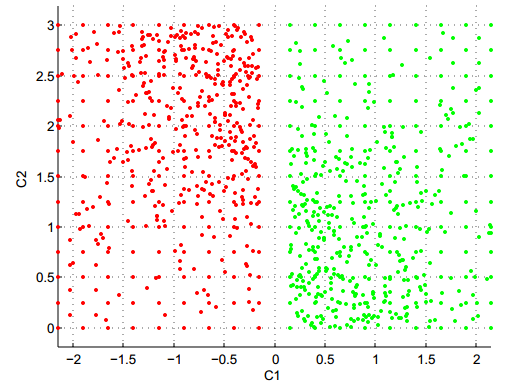
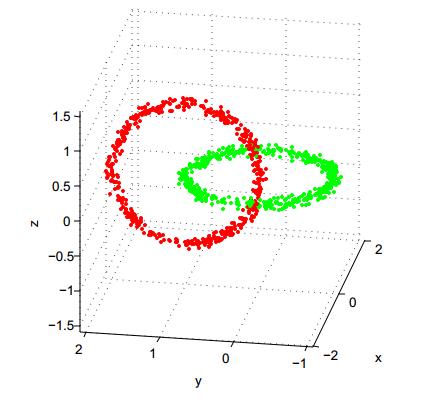
Now, experiment with the Hierarchical Agglomerative Clustering (HAC) methods (hclust function):
dist2_xxx=dist(true_data_xxx)
hs_xxx=hclust(dist2_xxx,method="single")
plot(hs_xxx) # For visualizing the DENDROGRAM
labS_xxx=cutree(hs_xxx,K)
# K = number of desired clusters, which shall determine the height at which the dendrogram will be cut
plot(true_data_xxx,col=labS_xxx)
hc_xxx=hclust(dist2_xxx,method="complete")
plot(hc_xxx) # To visualize the DENDROGRAM
labC_xxx=cutree(hc_xxx,K) # K = number of desired clusters, which shall determine the height at which the dendrogram will be cut
plot(true_data_xxx,col=labC_xxx)
QUESTION 3: On which dataset does "single-linkage" HAC produce an excellent result? Why is it logical?
QUESTION 4: Conversely, on which dataset does "single-linkage" HAC perform very badly, and why is it expected?
QUESTION 5: By comparing with what you had observed for K-means, which variant of HAC (between single-linkage and complete-linkage) seems to be the most complementary to K-means?